Entry: NAFlex_ProtDNA_1hlv
Nucleic Sequence:
| Sequence | AATCCCGTTTCCAACGAAGGC |
| Rev. Sequence | GCCTTCGTTGGAAACGGGATT |
| Type | Prot-DNA |
| SubType | B |
| Chains | duplex |
| Pdb | 1HLV |
| Ligands | No |
MD Simulation >> (Click to expand/shrink)
Unified Molecular Modeling (UMM) Metadata
(Click to see full UMM)
(Click to see full UMM)
| AdditionalIons | No |
| AdditionalMolecules | No |
| AdditionalSolvent | No |
| Author | P.D. |
| Chains | duplex |
| Comments | Prot-DNA study |
| CounterIons | Na+ |
| Format | netcdf |
| FrameStep | 2 ps |
| Frames | 1000000 |
| IonicConcentration | Electroneutrality |
| IonsParameters | Dang |
| Ligands | No |
| NucType | Prot-DNA |
| PDB | 1HLV |
| Parts | DNA |
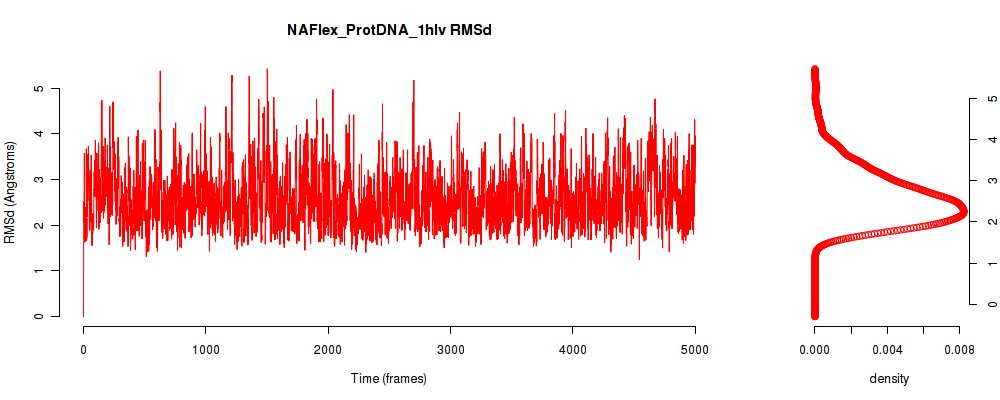
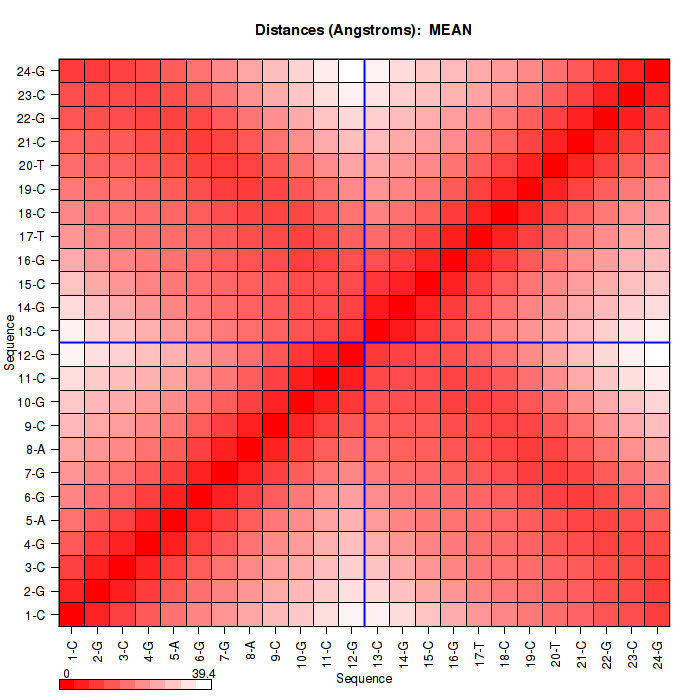
| RMSd_avg | 2.582 |
| RMSd_stdev | 0.618 |
| Rgyr_avg | 21.000 |
| Rgyr_stdev | 0.402 |
| SASA_avg | 7430.197 |
| SASA_stdev | 111.748 |
| SubType | B |
| Temperature | 300K |
| Topology | topol.top |
| Trajectory | traj.nc |
| Water | TIP3P |
| _id | NAFlex_ProtDNA_1hlv |
| boxAngle | 109.471219 |
| boxX | 86.2381964 |
| boxY | 86.2381964 |
| boxZ | 86.2381964 |
| dataset | Array |
| date | 0.00000000 1453420800 |
| description | DNA-B Duplex Naked ParmBSC1 TIP3P Electroneutral |
| forceField | parmBSC1 |
| ionsModel | - |
| moleculeType | Prot-Dna |
| rev_sequence | GCCTTCGTTGGAAACGGGATT |
| saltConcentration | - |
| sequence | AATCCCGTTTCCAACGAAGGC |
| soluteAtoms | 1331 |
| soluteCharge | -40 |
| soluteResidues | 42 |
| solventAtoms | - |
| solventResidues | - |
| time | 2,000 |
| totalAtoms | 1331 |
| totalCharge | -40 |
| totalIons | - |
| totalResidues | 42 |